|
 |
- Search
| Asian Spine J > Volume 11(4); 2017 > Article |
|
Abstract
Purpose
The etiology of Modic changes (MCs) is unclear. Recently, the role of genetic factors in the etiology of MCs has been evaluated. However, studies with a larger patient subset are lacking, and candidate genes involved in other disc degeneration phenotypes have not been evaluated. We studied the prevalence of MCs and genetic association of 41 candidate genes in a large Indian cohort.
Overview of Literature
MCs are vertebral endplate signal changes predominantly observed in the lumbar spine. A significant association between MCs and lumbar disc degeneration and nonspecific low back pain has been described, with the etiopathogenesis implicating various mechanical, infective, and biochemical factors.
Methods
We studied 809 patients using 1.5-T magnetic resonance imaging to determine the prevalence, patterns, distribution, and type of lumbar MCs. Genetic association analysis of 71 single nucleotide polymorphisms (SNPs) of 41 candidate genes was performed based on the presence or absence of MCs. SNPs were genotyped using the Sequenome platform, and an association test was performed using PLINK software.
Results
The mean age of the study population (n=809) was 36.7±10.8 years. Based on the presence of MCs, the cohort was divided into 702 controls and 107 cases (prevalence, 13%). MCs were more commonly present in the lower (149/251, 59.4%) than in the upper (102/251, 40.6%) endplates. L4–5 endplates were the most commonly affected levels (30.7%). Type 2 MCs were the most commonly observed pattern (n=206, 82%). The rs2228570 SNP of VDR (p=0.02) and rs17099008 SNP of MMP20 (p=0.03) were significantly associated with MCs.
Modic changes (MCs) are vertebral endplate signal changes observed in sagittal magnetic resonance imaging (MRI) sequences [1]. Although MCs have been described in the lumbar, thoracic, and cervical spine, they are predominantly observed in the lumbar spine. Three types of MCs have been described based on the signal changes in T1 and T2 sequences in MRI. Though the true significance of MCs is not known, a significant association between MCs and lumbar disc degeneration and nonspecific low back pain has been described in different studies [2345]. The prevalence of MCs varies from 18% to 62% in patients with low back pain, with types I and II being the most commonly observed patterns [23].
The etiopathogenesis of MCs is still not known, and various mechanical, infective, and biochemical reasons have been described. Albert et al. [6] postulated that with the presence of disc herniation and neovascularization, anaerobic bacteria can lodge in the endplates and initiate edema and inflammation. Elevated levels of inflammatory mediators, such as interleukins (IL) and proinflammatory cytokines, have been detected in endplates with MCs, suggesting an inflammatory cause [7]. With the initiation of disc degeneration, the increased stress on the endplates can result in microfractures and fissures within the endplates and this could be a major source of MCs [18].
As genetic factors have recently gained credence as possible etiologic mechanisms for disc degeneration, low back pain, and sciatica, the role of genetic factors in the etiology of MCs has also been recently evaluated by different researchers. Maatta et al. [9] studied 831 twins for heritability of MCs and concluded that MCs are heritable with 16%–43% prevalence of MCs heritability. Karppinen et al. [10] studied possible genetic associations of MCs in an occupational cohort of 159 male train engineers and 69 male paper mill workers. A total of 128 patients (56%) had MCs at one or more disc levels. None of the single nucleotide polymorphisms (SNPs) was significantly associated with MCs, but when gene–gene interactions were evaluated, IL-1A and matrix metalloproteinase 3 (MMP3) polymorphisms together were associated with type II MCs [10]. However, studies with a larger patient subset are lacking, and to the best of our knowledge, candidate genes involved in other disc degeneration phenotypes have not been studied. We, therefore, evaluated the potential role of genetic polymorphisms in 41 candidate genes (including 71 SNPs) with MCs in a larger cohort.
Institutional Review Board approval was obtained before starting this cross-sectional study. The study population was recruited from patients (Indian origin) presenting at the Spine Unit of Ganga Medical Centre and Hospital, Coimbatore, India. An individual was included in the study based on the following criteria: any sex, between 18 and 70 years, no evidence of other spinal diseases (congenital, pathologic, inflammatory, or infective), no history of previous lumbar spine surgery, and no history of significant spinal injury. Any patient who required MRI of the lumbar spine and who met the inclusion criteria was explained about the study and, if consent was provided, was included in the project.
All individuals were evaluated by MRI, including T1- and T2-weighted axial and sagittal sections of the lumbar spine (T12–S1) and a screening MRI T2-weighted sagittal image of the whole spine (1.5-Tesla MRI, Siemens, Munich, Germany). The scanning protocol was as follows: sagittal T1 turbo spine echo sequence of recovery time 400–750 milliseconds, echo time 10–13 milliseconds, slice thickness 4 mm, and field of view 360×360 mm and sagittal T2 turbo spin echo sequence of recovery time 2,800–3,400 milliseconds, echo time 50–100 milliseconds, slice thickness of 4 mm, and field of view of 360×360 mm. For analyzing MCs, only the lumbar region was studied. All five lumbar discs from L1 to S1 were analyzed, and in patients with transitional lumbosacral vertebra, the last five mobile discs were studied. The presence and type of MCs were assessed in the sagittal T1 and T2 sequences (Figs. 1, 2, 3). The presence of endplate signal changes in the anterior and posterior corners of the vertebral body near the disc and of signal changes subjacent to the entire endplate was considered, whereas signal changes adjacent to Schmorl's nodes were excluded. Five sagittal sequences, including one midsagittal and four parasagittal sections, were studied in all patients. The types of MCs were evaluated by two independent observers. The interobserver reliability between them was excellent (κ statistic 0.82±0.07). A genetic association analysis of all potential SNPs was performed with reference to the presence or absence of MCs (case vs. control analysis). Individuals without MCs in any of the lumbar discs were considered as controls, whereas those with MCs in either or both endplates at any one of the lumbar disc levels were considered as cases.
Blood samples were collected from the study population in ethylenediaminetetraacetic acid–containing tubes and stored at −80℃ for laboratory analysis. DNA was extracted from the frozen human blood. The quality and quantity of DNA was checked using agarose gel electrophoresis and spectrophotometry. A total of 71 SNPs in 41 candidate genes were selected for analysis based on previous genetic studies on lumbar disc degeneration (COL9A2, COL11A1, COL22A1, COL9A1, AGC1, SKT, CHST3, GLI1, CILP, CALM1, IGF1R, COMP, HAPLN1, FN1, BMP5, BMP2, HHIP, VDR, NGFB, TAC1, MMP2, MMP7, MMP20, MMP9, MMP10, MMP1, MMP3, MMP12, MMP13, ADAMTS5, ADH2, IL18RAP, IL1A, IL1B, IL1F5, IL6, IL1F10, IL1RN, COX2, LEPR, and ANP32A).
Genotyping of SNPs of case and control samples was performed using the Sequenome platform. SpectroAquire and MassARRAY Typer Software packages (Sequenom, San Diego, CA, USA) were used for interpretation and Typer analyser V3.4.0.18 was used to review and analyze all data.
PLINK was used for all the downstream analysis and validation data. SNPs prevalent in all target genes of the study population were predicted, and an association test was performed using PLINK software based on the nature of SNPs. Probability values and odds ratios were estimated for all 71 SNPs, and we also tested their association with MCs.
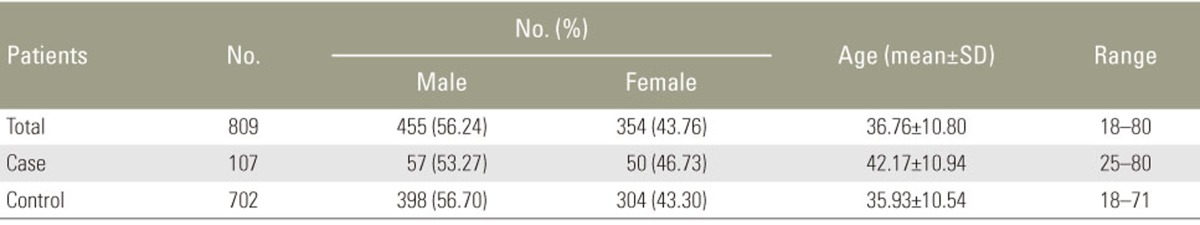
The 809 individuals who satisfied the inclusion criteria included 455 males and 354 females (Table 1). The mean age of the subjects was 36.7±10.8 years. Based on the presence of MCs at any one of the five lumbar discs, the total population was divided into 702 controls and 107 cases. There was no significant difference between the controls and cases in terms of age (35.9±10.5 vs. 42.2±10.9) and male:female distribution (56%:44% vs. 54%:46%).
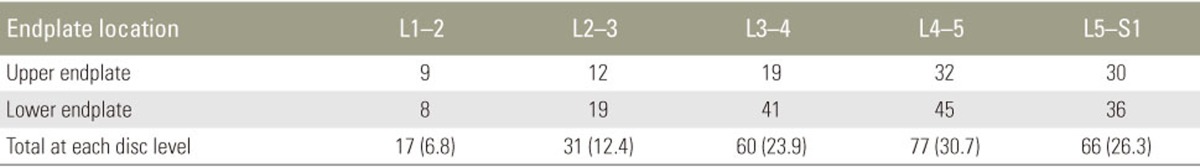
Among the 809 cases, 64 patients had single level (7.9%), 32 had double level (3.9%), five each had three and four level (0.6%), and one had multilevel MCs (0.1%). MCs were identified in 251 among the 1070 endplates. MCs were situated more commonly in the lower (149, 59.4%) than in the upper (102, 40.6%) endplates. The L4–5 endplates were the most commonly affected level (n=77, 30.7%) followed by L5–S1 (n=66, 26.3%), L3–4 (n=60, 23.9%), L2–3 (n=31, 12.4%), and L1–2 (n=17, 6.8%) (Table 2).
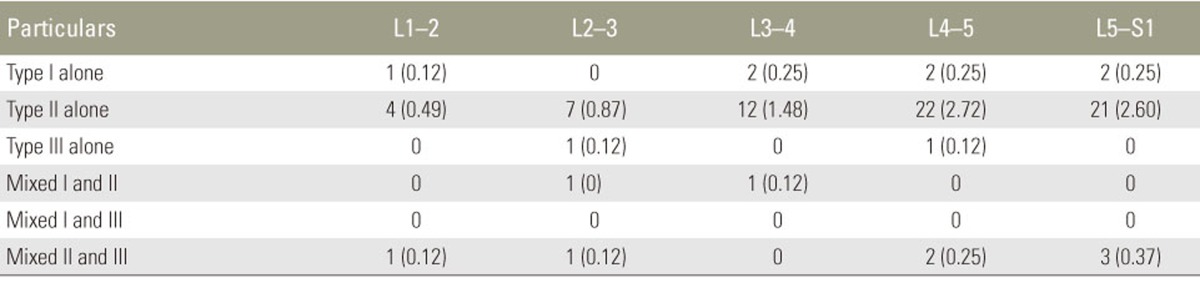
Among the 251 endplates with MCs, type 2 MCs were the most commonly observed pattern (n=206, 82%), followed by types 1 (n=27, 10.8%) and 3 (n=18, 7.2%). We analyzed the presence of mirroring endplate MCs on either side of a particular disc (i.e., the presence of the same type 1, 2, or 3 changes on both sides of a lumbar disc). Mirroring endplate MCs were observed in 75 of the total 535 discs in the 107 cases. Type 2 MCs were the most commonly observed pattern (n=66, 88%) followed by type 1 (n=7, 9.3%) and 3 (n=2, 2.7%) MCs. Mixed patterns were observed in nine discs including two mixed I and II and seven mixed II and III patterns. A mixed I and III pattern was not observed (Table 3).
Preliminary analysis of SNP genotyping data revealed that the success rate of genotyping (71 SNPs spread across 41 candidate genes genotyped in 809 individuals) was approximately98.5% and the SNP call rate was 91.83%. Association analysis between genotyping and phenotyping data using PLINK revealed a significant association between two SNPs, i.e., rs2228570 and rs17088008 with MC phenotype at p<0.05. One of thers2228570 SNPs located in a vitamin D receptor (VDR) gene showed greater degree of association (p=0.02) and the other rs17099008 SNP belonging to an MMP (MMP 20) gene had a p-value of 0.03 (Table 4).
Although de Roos et al. [5] described MCs for the first time, Modic et al. [1] classified these changes into three categories: type I changes showing low-signal intensity in T1-weighted images and high-signal intensity in T2-weighted images, type II changes showing high-signal intensity in T1 and T2 sequences, and type III changes showing low-signal intensity in T1 and T2 sequences. Type I changes are presumed to indicate an active inflammatory process in the subchondral region, whereas type II changes are presumed to reflect fatty degeneration of the bone marrow and type III changes are presumed to indicate sclerosis in the subchondral region.
The prevalence of MCs varies in different studies from 18% to 62% and the prevalence of each type also varies [23]. Based on previous studies, types I and II are the most common patterns in the lumbar spine. The inconsistent results regarding the prevalence of studies on MCs could be probably due to differences in number of patients, inclusion criteria, and study design. In their study on 59 asymptomatic subjects, Chung et al. [11] observed that type 2 changes (n=38) were more frequent than type I changes (n=11) in 590 lumbar vertebral endplates. In our study also, type 2 MCs were the most commonly observed pattern (n=206, 82%). Types 1 (10.8%) and 3 (7.2%) were very infrequent. Among the different lumbar levels, Modic et al. [1] observed that the distribution of MCs at L4–L5 or L5–S1 was the most common. These observations were confirmed by Kuisma et al. [12]. They noted that the distribution of types I and II was higher in the anterior third of the vertebra than in the posterior two-thirds of the vertebra and the distribution of type II was predominant in the superior endplate versus that in the inferior endplate [12]. In the present study, MCs were more commonly situated in the lower (149, 59.4%) than in the upper (102, 40.6%) endplates. The L4–5 endplates were the most commonly affected level (77, 30.7%) followed by L5–S1 (26.3%), L3–4 (23.9%), L2–3 (12.4%), and L1–2 (6.8%).
Pathophysiologically, MCs have been considered to arise following biomechanical and biochemical insults to the endplate. With progressive lumbar disc degeneration, increased stress on the endplates causes calcification and microfractures, which result in neovascularization, edema, and inflammatory response [1]. This is observed as type 1 MCs. Altered endplate function interferes with the nutrition to the discs perpetuating further degeneration of the nucleus pulposus. Modic et al. [14] have demonstrated that type I MCs are characterized by disruption of and fissures in the endplates. Once the acute inflammation settles, the inflammatory tissue is replaced by fatty marrow in the subchondral region, which is observed as hyperintense signal changes in T1 and T2 sequences (type II MCs). In chronic situations, sclerosis and calcification in the subchondral marrow are visualized as type III MCs. Apart from biomechanical stress induced by the degenerated discs on the endplate, active inflammation induced by inflammatory cells, ILs, and cytokines seems to have a large role in the pathogenesis of MCs [36]. Using the immunohistochemical method, Ohtori et al. [13] found that protein gene product 9.5–immunoreactive nerve fibers and tumor necrosis factor (TNF)-immunoreactive cells in the endplates from patients with MCs were significantly greater than those in the endplates from normal subjects on MRI.
In the present study, among the 71 SNPs studied, two SNPs (VDR and MMP20) had a significant association with the presence of MCs. Two previous studies on the genetic association of MCs have observed different genes to be implicated, including IL1 cluster, MMP3, ADAMTS, and TNF-α. Karppinen et al. [10] studied eight genes in an occupational cohort of 159 male train engineers and 69 male paper mill workers. They specifically studied type II MCs and observed that none of the SNPs was significantly associated with MCs when analyzed independently, but when gene-gene interactions were evaluated, IL1A and IL1 gene cluster together with MMP3 polymorphisms were associated [10]. In the present study, MMP20 had a significant association. Though IL1 and MMP3 polymorphisms were studied, we could not derive any association. In the study by Karppinen et al. [10], of the 228 subjects, 128 (56%) had MCs at one or more disc levels. In our study, 107 subjects had MCs, the prevalence being 13% of 809 individuals. The wide age range, inclusion criteria, and variable occupation profile of the study subjects could be the reasons for the lesser incidence in the present study. In a subsequent study by the same authors, but on a patient group from a different geographical area, among 108 men from three different occupational groups, the presence of the minor allele of IL1A was associated with MCs [9]. Though both studies underline the importance of the IL1A gene in the pathophysiology of MCs, we did not observe an association with any of the IL1 genes (IL18RAP, IL1A, IL1B, IL1F5, IL1F10, and IL1RN) in our study.
The rs2228570 SNP of the VDR gene had a strong association (p=0.02) with the presence of MCs in our study. VDR is a steroid receptor and has an important role in calcium homeostasis, normal bone mineralization, and remodeling. VDR was the first reported gene associated with disc degeneration in a study onmonozygotic twins in a Finnish population [14] and later confirmed in two other studies [1516]. In a report of 342 individuals from an Indian cohort, Rajasekaran et al. [17] studied the genetic association of 58 SNPs with different MRI parameters of disc degeneration, including Pfirrmann's grading, annular tear, disc bulge, Schmorl's nodes, and endplate damage score. Though 11 SNPs provided evidence of association with one of the phenotypes, the frequency of the risk t-allele of VDR was very low in this study and an association could not be found.
The rs17099008 SNP of MMP (MMP20, p=0.03) was significantly associated with MCs. MMPs are a group of enzymes that participate in the degradation of the major structural components of the intervertebral disc and are known to be involved in the normal turnover and pathologic degradation of the extracellular matrix in the connective tissue. Though other MMPs, like MMP3 and MMP10, have been linked to disc degeneration and MCs, the association of MMP20 with MCs has been observed for the first time. In the study by Karppinen et al. [10], it was observed that the combination of the T-C haplotype of IL1A and the MMP3 minor 5A allele was associated with eight-fold odds on MCs among middle-aged Finnish men.
MCs are vertebral endplate changes observed in approximately 13% of the present study population. The significance and etiology of these changes are not clear but have been implicated increasingly with low back pain. The present study identifies genetic polymorphisms of VDR and MMP20 to be significantly associated with MCs in our study population. To the best of our knowledge, these associations have not been reported previously. Understanding the etiopathogenetic mechanisms of MCs could help us plan preventive and therapeutic strategies.
Acknowledgments
The study is funded by Ganga Orthopaedic Research and Education Foundation and ASSI research award 2014.
This study was approved by Institutional Review Board of Ganga Hospital, Coimbatore, India.
Notes
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
References
1. Modic MT, Steinberg PM, Ross JS, Masaryk TJ, Carter JR. Degenerative disk disease: assessment of changes in vertebral body marrow with MR imaging. Radiology 1988;166(1 Pt 1): 193–199. PMID: 3336678.


2. Albert HB, Manniche C. Modic changes following lumbar disc herniation. Eur Spine J 2007;16:977–982. PMID: 17334791.



3. Kjaer P, Korsholm L, Bendix T, Sorensen JS, Leboeuf-Yde C. Modic changes and their associations with clinical findings. Eur Spine J 2006;15:1312–1319. PMID: 16896838.



4. Modic MT, Masaryk TJ, Ross JS, Carter JR. Imaging of degenerative disk disease. Radiology 1988;168:177–186. PMID: 3289089.


5. de Roos A, Kressel H, Spritzer C, Dalinka M. MR imaging of marrow changes adjacent to end plates in degenerative lumbar disk disease. AJR Am J Roentgenol 1987;149:531–534. PMID: 3497539.


6. Albert HB, Kjaer P, Jensen TS, Sorensen JS, Bendix T, Manniche C. Modic changes, possible causes and relation to low back pain. Med Hypotheses 2008;70:361–368. PMID: 17624684.


7. Burke JG, Watson RW, McCormack D, Dowling FE, Walsh MG, Fitzpatrick JM. Intervertebral discs which cause low back pain secrete high levels of proinflammatory mediators. J Bone Joint Surg Br 2002;84:196–201. PMID: 11924650.


8. Schmid G, Witteler A, Willburger R, Kuhnen C, Jergas M, Koester O. Lumbar disk herniation: correlation of histologic findings with marrow signal intensity changes in vertebral endplates at MR imaging. Radiology 2004;231:352–358. PMID: 15064391.


9. Maatta JH, Kraatari M, Wolber L, et al. Vertebral endplate change as a feature of intervertebral disc degeneration: a heritability study. Eur Spine J 2014;23:1856–1862. PMID: 24828957.


10. Karppinen J, Daavittila I, Solovieva S, et al. Genetic factors are associated with modic changes in endplates of lumbar vertebral bodies. Spine (Phila Pa 1976) 2008;33:1236–1241. PMID: 18469698.


11. Chung CB, Vande Berg BC, Tavernier T, et al. End plate marrow changes in the asymptomatic lumbosacral spine: frequency, distribution and correlation with age and degenerative changes. Skeletal Radiol 2004;33:399–404. PMID: 15138721.


12. Kuisma M, Karppinen J, Niinimaki J, et al. Modic changes in endplates of lumbar vertebral bodies: prevalence and association with low back and sciatic pain among middle-aged male workers. Spine (Phila Pa 1976) 2007;32:1116–1122. PMID: 17471095.


13. Ohtori S, Inoue G, Ito T, et al. Tumor necrosis factorimmunoreactive cells and PGP 9.5-immunoreactive nerve fibers in vertebral endplates of patients with discogenic low back pain and Modic Type 1 or Type 2 changes on MRI. Spine (Phila Pa 1976) 2006;31:1026–1031. PMID: 16641780.


14. Videman T, Leppavuori J, Kaprio J, et al. Intragenic polymorphisms of the vitamin D receptor gene associated with intervertebral disc degeneration. Spine (Phila Pa 1976) 1998;23:2477–2485. PMID: 9854746.


15. Kawaguchi Y, Kanamori M, Ishihara H, Ohmori K, Matsui H, Kimura T. The association of lumbar disc disease with vitamin-D receptor gene polymorphism. J Bone Joint Surg Am 2002;84:2022–2028. PMID: 12429765.


16. Cheung KM, Chan D, Karppinen J, et al. Association of the Taq I allele in vitamin D receptor with degenerative disc disease and disc bulge in a Chinese population. Spine (Phila Pa 1976) 2006;31:1143–1148. PMID: 16648751.


17. Rajasekaran S, Kanna RM, Senthil N, et al. Phenotype variations affect genetic association studies of degenerative disc disease: conclusions of analysis of genetic association of 58 single nucleotide polymorphisms with highly specific phenotypes for disc degeneration in 332 subjects. Spine J 2013;13:1309–1320. PMID: 23792102.


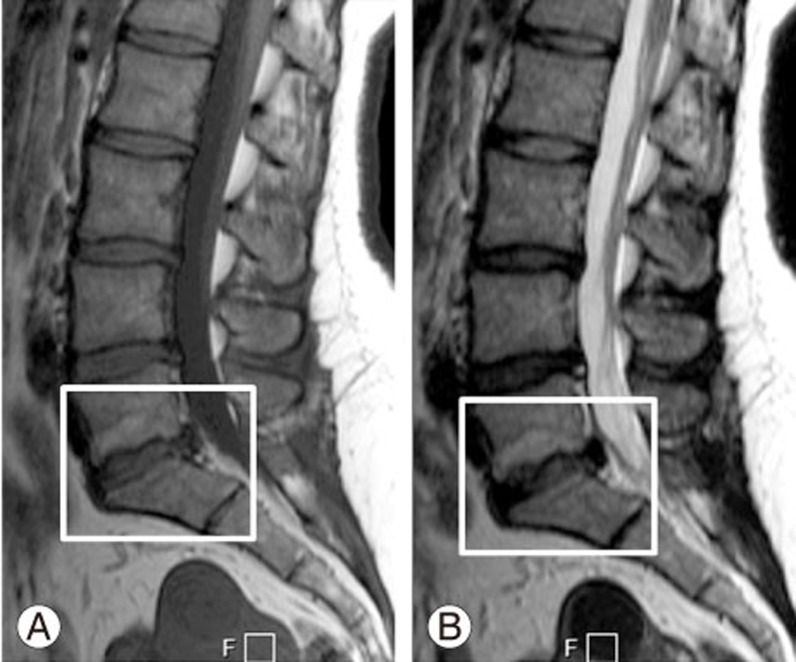
Fig. 1
Type 1 Modic changes. (A, B) Hypointense T1 signal and hyperintense T2 signal changes are observed in the L3–4 lower endplate.
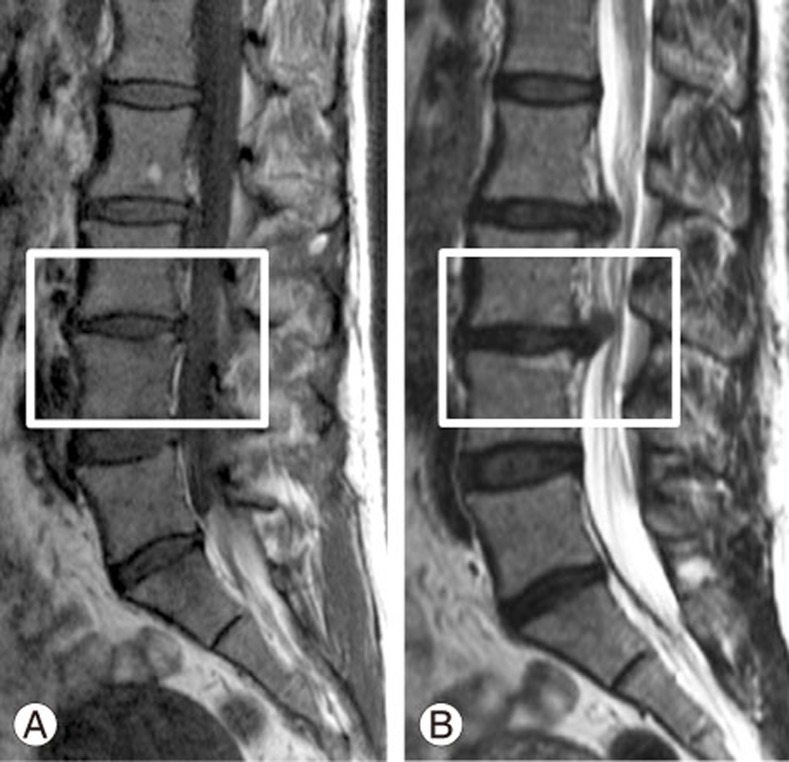
Fig. 2
Type 2 Modic changes. (A, B) Hyperintense T1 signal and T2 signal changes are observed in the L5–S1 superior endplate.
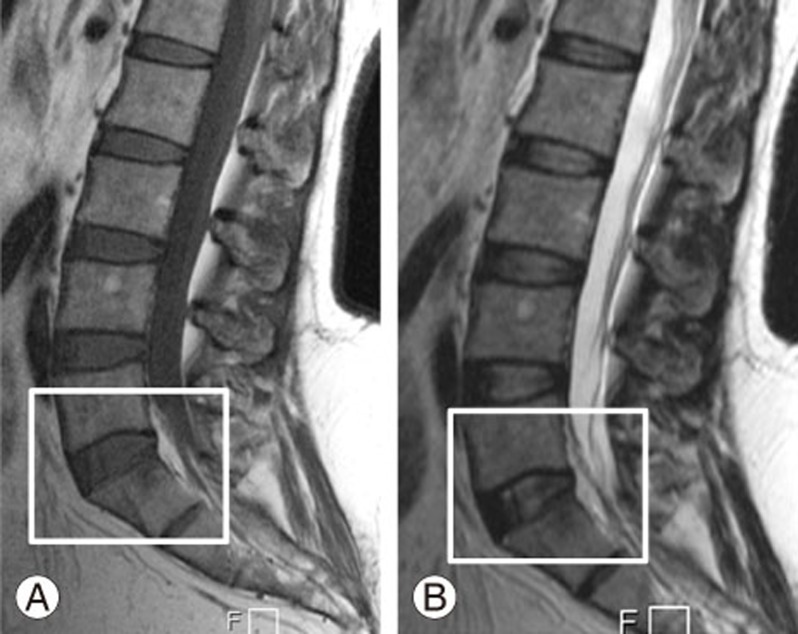
Fig. 3
Type 3 Modic changes. (A, B) Hypointense T1 signal andT2 signal changes are observed in the L5–S1 inferior endplate.