Introduction
Tumor endothelial markers (TEMs) are a group of cell surface proteins specifically expressed in the endothelium of various cancers, but are not expressed in normal endothelial tissues1-3. The limited expression profile of TEMs in the tumor endothelium indicates specific and restricted functions of the proteins in tumor angiogenesis2. TEMs associated with the cell surface are particularly of interest in that cell surface targets are directly accessible via the blood stream for functional intervention.
Several recent studies have showed evidence implicating a role of TEMs in normal tissues. For example, TEM1 is the lectin-like cell surface protein endosialin that is normally expressed in several peripheral tissues2, and TEM8 has been recently identified as the receptor for anthrax toxin4. It is also known that the mouse homologues of TEM1, TEM5 and TEM8 are abundantly expressed in the endothelium of the embryonic liver2. Therefore, it appears that TEMs participate in a normal physiological process as well as in tumor angiogenesis.
Among the uncharacterized TEMs, TEM7 is a single transmembrane protein that contains the plexin-semaphorin-integrin (PSI) domain in its presumptive extracellular region2. The PSI domain is mainly found in cell surface proteins of the nervous system, such as semaphorin and its receptor, plexin. The semaphorin family is a major repellent that regulates axon guidance and neuronal migration in the developing nervous system5,6. Interestingly, these neuronal guidance molecules having a PSI domain play an important role outside the nervous system, especially in the vascular system. For example, it was recently reported that semaphorin-3F and its receptor, neuropilin, regulate angiogenesis in vitro and in vivo7-10. Reversely, certain angiogenic factors appear to have an important function in the nervous system. For example, VEGF is known to regulate neuronal cell migration, neurite maturation and neurogenesis11-13. In terms of TEM7, Carson-Walter et al.2, recently demonstrated the presence of TEM7 mRNA in cerebellar Purkinje cells of the mouse brain, implicating a possible role of the endothelial protein, TEM7, in the nervous system. In the present study, to determine the expression profile and function of TEM7 in the spinal nerves, the histochemical localization of TEM7 in the spinal cord and dorsal root ganglion (DRG) of developing rats and adult rats was investigated.
The PSI domain, which was originally considered as a part of the sema domain, is found in several transmembrane proteins14. Even though the presumptive function of the PSI domain is protein binding14,15, biological evidence for a function of the PSI domain is still lacking. Most PSI proteins are type I transmembrane proteins that function as receptors or secreted ligands. The predicted structure of TEM7 suggests that it is also a type I transmembrane protein with a large extracellular region (ectodomain), a hydrophobic transmembrane domain, and a short cytoplasmic tail2. A secreted form of the ectodomain of type I transmembrane proteins fused with a tag has been created to localize the expression of the corresponding ligands of the transmembrane proteins16. The present study employed the same strategy to analyze the expression profile of a putative ligand of TEM7 in the spinal cord and DRG using a recombinant TEM7 ectodomain.
Materials and Methods
1. cRNA probes and in situ hybridization immunohistochemistry
For producing a TEM7 cRNA probe, a cDNA fragment of rat TEM7 was used as a template. The plasmid containing the fragment of rat TEM7 cDNA was linearized at NcoI or SalI sites located in the polylinker for antisense and sense probes, respectively.
Labeling of RNA probes with digoxigenin-11-UTP (DIG-11-UTP) was performed using an in vitro transcription kit (Roche Diagnostics). The amount and size of the transcribed cRNA were estimated with RNA gel electrophoresis.
In situ hybridization was performed as previously described with minor modifications17,18. Briefly, frozen spinal cords of adult Sprague-Dawley rats (p50~70, n=10) were sectioned at 18 ┬Ąm thicknesses on a cryostat (Leica). For developmental analysis, Sprague-Dawley rat pups (n=12 in total) were sacrificed at postnatal day 3, 10, and 20. The spinal cord and DRG were carefully dissected under a stereomicroscope (Olympus), and the central and peripheral branches of the DRG were removed with a fine scissor. The damaged DRG was discarded, and the intact DRG and spinal cord were frozen with dry ice. Frozen sections were prepared with the cryostat, and the sections were fixed in 4% paraformaldehyde (PFA) for 10 minutes, washed three times with phosphate buffered saline (PBS), and finally acetylated for 10 minutes. After prehybridization, the sections were incubated with hybridization buffer (50% formamide, 4├ŚSSC, 0.1% CHAPS, 5 mM Na2EDTA, 0.1% Tween-20, 1.25├ŚDenhartdt's, 125 ┬Ąg/mL yeast tRNA, 50 ┬Ąg/mL heparin, 200 ng digoxigenin-labeled probes) for 18 hours at 57Ōäā. Non-specific hybridized proteins were removed by washing in 2├ŚSSC for 10 minutes and with RNase A (10 ┬Ąg/mL), treatment), followed by a final washing step in 0.1├ŚSSC at 57Ōäā for 15 minutes. For immunological detection of digoxigenin-labeled hybrids, the sections were incubated with anti-digoxigenin alkaline phosphatase (1:1500; Roche Diagnostics) for 1 hour, and the color reaction was carried out with 4-nitroblue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP). Sections were dehydrated and were mounted with Crystalmount.
2. Molecular cloning of the TEM7 ectodomain
The ectodomain of human TEM7 (24aa-412aa) was generated using the polymerase chain reaction (PCR) with a human TEM7 cDNA template (a gift from Dr. B. Vogelstein). The sense PCR primers contained an SfiI site whereas the antisense primers had HindIII sites. After restriction enzyme digestion, the PCR fragments were inserted into the AP5 vector (a gift from Dr. Z. He) which contains alkaline phosphatase (AP), Myc and His as expression tags.
3. Expression and purification of the TEM7 ectodomain
HEK 293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics. The cells were transfected with 10 ┬Ąg of the expression vectors by the calcium phosphate method. The conditioned medium and transfected cells were harvested two days after transfection, and were solubilized with 2├Śsodium dodecyl sulfate (SDS) loading dye. The samples were subjected to SDS polyacrylamide gel electrophoresis and were transferred onto a nitrocellulose membrane (Hybond-ECL, Amersham). After blocking in 5% non-fat milk in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST), the same membranes were incubated with anti-His or anti-Myc antibodies overnight at 4Ōäā. Next, the membranes were incubated with horseradish peroxidase conjugated anti-mouse or anti-rabbit IgG (1:3000; Amersham) for 1 h at room temperature. An enhanced chemiluminescence reaction (Amersham) was performed to visualize the reaction, and the blot was analyzed using a luminescent image analyzer (LAS-3000; Science Lab).
Individual AP fusion proteins were purified with nickel-nitrilotriacetic acid beads. Conditioned medium from the transfected HEK 293 cells was passed over the nickel charged resins, and unbound proteins were eluted with buffer B (500 mM NaCl, 20 mM Tris-HCl, 5 mM imidazole, pH 7.9) and buffer W (500 mM NaCl, 20 mM Tris-HCl, 60 mM imidazole, pH 7.9). The AP fusion proteins were recovered by elution with buffer E (500 mM NaCl, 20 mM Tris-HCl, 1M imidazole, pH 7.9), and were concentrated using Vivaspin concentrators (Viva science). SDS-PAGE and Coomassie blue staining confirmed purity (>90%).
4. Measurement of alkaline phosphatase activity
To measure of the activity of tagged AP of the fusion protein, conditioned media from stably transfected cells or from control HEK cells were used. The conditioned media were heated at 65Ōäā for 10 minutes to inactivate the endogenous AP activity, and were then centrifuged at 13,000 rpm for 5 minutes. The supernatant was incubated with 200 ┬ĄL of 1├ŚAP buffer (1.65 mg/mL p-nitro phenyl phosphate, 0.5 mM MgCl2, 0.1 M diethanolamine (pH 9.8) for the indicated time, and the amount of color reaction was measured using a spectrophotometer at a wavelength of 405 nm.
5. Immunohistochemistry
Adult male Sprague-Dawley (p50~70, n=20) were used for immunohistochemical analysis and the frozen ligand binding assay. All procedures were performed according to protocols approved by the Dong-A University Committee on animal research that follow the guidelines of animal experimentation established by The Korean Academy of Medical Sciences. Animals were deeply anesthetized with an intraperitoneal injection of phentobarbital and were perfused with ice-cold PBS (pH 7.2), followed by 4% PFA in PBS. The brains, spinal cord and dorsal root ganglia were removed and cryoprotected with 30% sucrose overnight and cryostat sections (30 ┬Ąm) were subsequently prepared. Tissue sections were permeated with 0.2% Triton X-100 in PBS for 15 minutes. After blocking with 10% normal goat serum for 1 hour at room temperature, the sections were incubated with polyclonal anti-TEM7 antibody (1:2500) overnight at 4Ōäā. Sections were washed three times with PBS, and then sections were incubated with biotinylated anti-rabbit IgG (1:200; Vector Laboratories) for 2 h at room temperature. The sections were washed three times with PBS and were incubated with a mixture of avidin-biotin in PBS (1:100) for 1 h at room temperature. The chromogenic reaction was carried out with 0.05% diaminobenzidine and 0.03% hydrogen peroxide in 0.05 M Tris-Cl buffer (pH 7.4). Sections were dehydrated through a series of graded alcohol, cleared with xylene, and mounted with Crystalmount.
6. Ligand binding assay in tissue sections
Frozen sections were fixed in 4% PFA in PBS for 10 minutes at room temperature, washed twice in PBS for 5 minutes, and were then rinsed in ice-cold Hank's balanced salt solution (HBSS) containing 20 mM hepes for 10 minutes. The sections were incubated with conditioned medium containing TEM7-AP or AP for 1 hour at room temperature, washed twice in HBSS for 10 minutes, and then fixed in AP fix solution (65% acetone, 3% formaldehyde, 20 mM Hepes, pH 7.5). Next, the sections were heated in PBS for 2 hours at 65Ōäā, rinsed in AP buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 5 mM MgCl2) for 10 minutes to inactivate the endogenous alkaline phosphatase. Sections were developed with a developing solution containing NBT and BCIP for 5~10 minutes. Sections were then dehydrated through an ethanol series (70, 90, and 100% ethanol for 2 minutes each) and cleared twice in xylene. The sections were mounted with Crystalmount and were analyzed using a Zeiss Axiophot microscope.
Results
1. Localization of TEM7 mRNA in the spinal cord and dorsal root ganglion
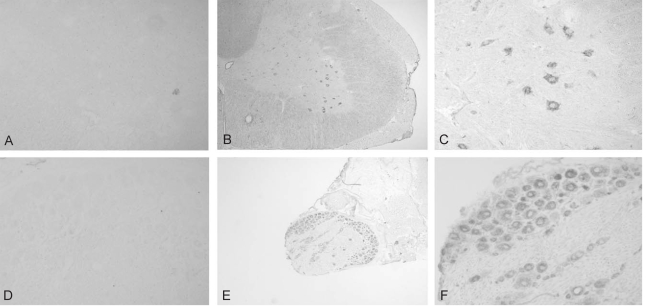
The expression of TEM7 mRNA in the spinal cord and DRG was analyzed with in situ hybridization using a digoxigenin-labeled antisense riboprobe. In the spinal cord, the antisense probe specifically hybridized with TEM7 mRNA in the neuronal population of the gray matter (Fig. 1B and 1C), whereas the sense probe did not hybridize with TEM7 mRNA (Fig. 1A). The specific signals were mainly observed in the motor neurons of the ventral horn, but other neurons in the gray matter, including the dorsal horn, showed a very low level of TEM7 mRNA expression. The hybridization signals were observed only in the cytoplasm but not in the nuclei of the motor neurons (Fig. 1C). The expression of TEM7 mRNA in the motor neurons was observed with the same pattern throughout the whole length of the spinal cord. There were no specific hybridization signals in the white matter of the spinal cord, meninges and blood vessels.
In the DRG, TEM7 mRNA was strongly expressed in the cytoplasm of sensory neurons, but the nuclei of the sensory neurons were devoid of hybridization signals (Fig. 1E and 1F). The expression of TEM7 mRNA in the DRG was found in cells with various sizes, implicating nonselective TEM7 expression in various sensory neurons in the DRG. The hybridization signals in the satellite cells, Schwann cells and the spinal nerves were negative. The analysis of the sections hybridized with the sense probe showed no specific signals in the DRG (Fig. 1D).
2. Cloning and characterization of the TEM7 ectodomain
The predicted ectodomain of human TEM7 (eTEM7) (Fig. 2A) was forcefully expressed in HEK 293 cells. The expression of eTEM7, which is expected to secrete a ~110 KDa protein composed of a TEM7 ectodomain, alkaline phosphatase (AP; 67 KDa) (Fig. 2A) and Myc/His tags, occurred not only in cell pellets but also in the culture medium of transiently transfected cells (Fig. 2B). This indicates that TEM7 is a type I transmembrane protein that has a single extracellular domain with the N-terminal outside of the cell. The transfection of the empty vector (AP-5), which contains AP as a tag, also produced AP, and AP was observed both in the cell pellet and supernatant (Fig. 2B).
The activity of the tagged AP of TEM7 ectodomain was analyzed to demonstrate the utility of this fusion protein for localizing the expression of the TEM7 ligand. The controlled medium or purified TEM7 proteins were used to test AP activity. AP and TEM7-AP exhibited high AP activity as compared with the controlled HEK medium, and the activity was dose-dependent (Fig. 2C). This finding indicates that the secreted fusion protein is functionally active and that the construct could be a useful tool for analyzing the localization profile of the TEM7 ligand.
3. Immunohistochemical analysis of TEM7 expression in the spinal cord and dorsal root ganglion
A polyclonal anti-TEM7 antibody that was purified with an affinity column was used to demonstrate the expression of TEM7 in the spinal cord and DRG at the protein level. The cerebellar section was used as a positive control for immunohistochemical analysis since TEM7 mRNA is known to be highly expressed in the cerebellar Purkinje cells2. Consistent with the results of in situ hybridization, the antibody specifically labeled the cerebellar Purkinje cells, but not other cells such as granule cells (Fig. 3B). However, with the omission of the primary antibody, no cellular structures were labeled (Fig. 3A).
Next, immunohitochemical analysis of TEM7 protein expression in the spinal cord and DRG was investigated. The TEM7 antibody labeled neuronal cells especially in the ventral horn of the spinal cord (Fig. 3C and D). In the white matter of the spinal cord, the axoplasm seemed to show weak immunoreactivity. In the DRG, the anti-TEM7 antibody specifically labeled the sensory neurons and the immunoreactivity appeared in the cytoplasm, but not in the nuclei (Fig. 3E and F). The finding that the expression of TEM7 protein observed in almost all sensory neurons is consistent with the finding of TEM7 mRNA expression.
4. Localization of a putative ligand of TEM7 in the spinal cord and dorsal root ganglion
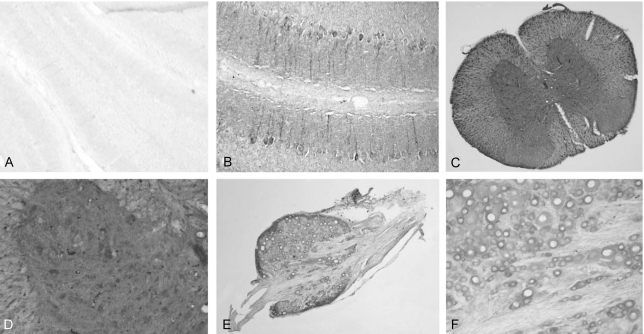
The expression of the putative ligand of TEM7 was analyzed by staining the binding site of the recombinant eTEM7-AP protein. In the spinal cord, the highest eTEM-AP binding site was observed in the dorsal and ventral root of the spinal nerves (Fig. 4A). The binding seemed to be observed in the neurilemma and endoneurium, but not in the axoplasm, axolemma and myelin sheath. The perineurium that surrounds the nerve fascicle also showed the strongest eTEM7-AP binding. In the gray matter, a very low level of eTEM7-AP binding was observed (Fig. 4A), but the cell bodies of the motor neurons which expressed TEM7 mRNA did not show any binding of eTEM7-AP. There was no labeling of eTEM7-AP in the neural structures of the white matter such as the axon fibers, myelin sheath, and extracellular matrix (Fig. 4B). However, specific binding of eTEM7-AP is clearly found in the wall of blood vessels (Fig. 4B). As a negative control, AP protein was used to stain the sections of the spinal cord. AP did not bind any structures in the spinal cord (data not shown).
In accordance with the strong reactivity of the neurilemmal structure of the spinal nerve roots to eTEM7-AP, the spinal nerves within the DRG also showed strong eTEM7- AP binding (Fig. 4C and D). In addition, the plasma membrane of sensory neurons in the DRG had prominent eTEM7-AP binding activity (Fig. 4C).
5. Developmental expression of TEM7 mRNA and its putative ligand in the spinal cord and dorsal root ganglion
Using in situ hybridization, the developmental changes of TEM7 mRNA expression in the spinal cord and DRG were analyzed. TEM7 mRNA in the motor neurons of the spinal cord first appeared on postnatal day 3, but the expression level was very low (Fig. 5A). A significant increase of TEM7 mRNA was observed during second postnatal weeks (Fig. 5B and C). However, the glial cells and endothelial cells did not show expression of TEM7 mRNA throughout the postnatal developmental period. In the DRG, expression of TEM7 mRNA was not observed at postnatal day 3 and 10 (Fig. 5D and E). TEM7 mRNA expression in the sensory neurons in the DRG was obviously found at postnatal day 20, and expression was confined to the sensory neurons, and not in other structures (Fig. 5F).
The ligand binding assay showed that the TEM7 binding activity of the spinal nerves appeared as early as postnatal day 3 (Fig. 6A). The binding was mainly observed in the neurilemma of the nerve roots with a similar binding pattern of eTEM7-AP in the adult spinal cord. The intensity of the staining in the spinal nerve root did not significantly change with age (Fig. 6B). The gray matter showed a low level of ligand binding, but the white matter did not, during the postnatal period (Fig. 6A and B).
Discussion
The present study is the first to show neuronal expression of TEM7 mRNA and protein in the spinal cord and DRG of rats. TEM7 mRNA expression was not observed in ependymal cells and endothelial cells of the spinal cord and DRG. mRNA expression in spinal neurons was demonstrated by immunohistochemical staining of TEM7 protein in motor and sensory neurons in the spinal cord and DRG, respectively. These findings indicate that TEM7 may play a specific role in several neuronal cells of the spinal nervous system.
The predicted structure of TEM7 is that of a type I transmembrane protein with a large extracellular region (ectodomain), a hydrophobic transmembrane domain, and a short cytoplasmic tail2. A secreted form of the ectodomain of type I transmembrane proteins that was fused with a tag has been made to localize the expression of the corresponding ligands of the ectodomains16. The present study employed the same strategy to make a secreted form of TEM7 ectodomain, and the localization of the putative ligand in the spinal cord and DRG was investigated using a recombinant TEM7 ectodomain. We found that the coverings of spinal nerves, such as the neurilemma, endoneurium and perineurium, show strong ligand binding to the TEM7 ectodomain. In the DRG, the ligand binding appeared around the sensory neurons and perineural structures of the spinal nerves. This finding indicates that the putative ligand of TEM7 may be a perineurial extracellular matrix (ECM) protein or a secreted extracellular protein.
The PSI domain consists of ~50 residues of amino acids and usually contains eight cysteine residues14. The cysteine residues are expected to be disulfide bonded. Even though the presumptive function of the PSI domain is protein-binding4,15, the biological function of the PSI domain is still unknown. Most PSI proteins are highly expressed during the developmental period in the nervous system, suggesting a role of the PSI domain-containing protein in the developmental processes such as axon guidance and neuronal migration. These developmental phenomena are tightly regulated by the interaction between the cell membrane receptor and ECM proteins. Therefore, the molecular structure of TEM7 and the localization profile of the putative TEM7 ligand suggest a possible role of TEM7 in neural development, especially in the spinal cord and DRG.
The majority of ECM proteins are structural proteins such as laminin and fibronectin19, and ECM proteins play a critical role in the development of various tissues. In the nervous system, ECM proteins regulate the adhesion and migration of growth cones and neuronal cells during development20. The present study indicates that the TEM7 ligand may be a perineurial ECM molecule. In contrast to the strong staining of TEM7 ligand in the perineural structure of peripheral spinal nerves, there was no staining in the white matter of the spinal cord. This finding indicates that the putative TEM7 ligand is expressed in certain cells of the peripheral nervous system. It may be possible that Schwann cells that surround the peripheral nerves will produce the putative TEM7 ligand. As the basal lamina of Schwann cells provide important substrates not only for developing axons but also for regenerating axons21, the putative TEM7/ECM ligand interaction may play a role in neurite growth during development and/or regeneration.
The present study has also revealed that the expression of TEM7 mRNA in the spinal cord and DRG was developmentally regulated. The expression increased during postnatal development. In addition, the ligand binding assay showed expression of the putative ligand of TEM7 as early as postnatal day 3 in the spinal nerve fibers. This finding indicates that TEM7-ligand interaction plays a role in the postnatal differentiation of spinal motor neurons and sensory neurons of DRG. TEM7 mRNA expression in the Purkinje cells of the cerebellum also increased during postnatal development (preliminary observation). Therefore, it seems that TEM7 might play an important role in the postnatal development of vertebrate neurons. Further studies on the function of TEM7 and its ligand in spinal nerve development would provide an insight into a novel mechanism of spinal cord development.
Even though the present and previous studies have revealed the expression profile of TEM7 in the nervous system and tumor endothelial cells, the function of TEM7 in neurons and endothelial cells is completely unknown at present. TEM7 seems to be involved in tumor angiogenesis in concert with other TEMs. It has been recently reported that TEM8 binds to an ECM protein, collagen alpha 322. The present study has revealed a possibility of an ECM protein as a TEM7 ligand. It was also reported that the TEM7 ectodomain inhibits fibrin-induced angiogenesis in vitro23, indicating a possible interaction between the TEM7 ectodomain and ECM matrix may regulate angiogenesis. Thus, the identification of TEM7 ligand will shed new light on the elucidation of TEM7 function in the nervous system as well as in tumor angiogenesis.